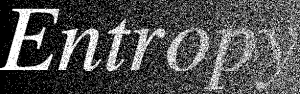Archive for the ‘Open Data Science’ Category
Feel free to discuss and contribute to this article over at the corresponding GitHub repo.
Many people suggest that you should use version control as part of your scientifc workflow. This is usually quickly followed up by recommendations to learn git and to put your project on GitHub. Learning and doing all of this for the first time takes a lot of effort. Alongside all of the recommendations to learn these technologies are horror stories telling how difficult it can be and memes saying that no one really knows what they are doing!
There are a lot of reasons to not embrace the git but there are even more to go ahead and do it. This is an attempt to convince you that it’s all going to be worth it alongside a bunch of resources that make it easy to get started and academic papers discussing the issues that version control can help resolve.
This document will not address how to do version control but will instead try to answer the questions what you can do with it and why you should bother. It was inspired by a conversation on twitter.
Improvements to individual workflow
Ways that git and GitHub can help your personal computational workflow – even if your project is just one or two files and you are the only person working on it.
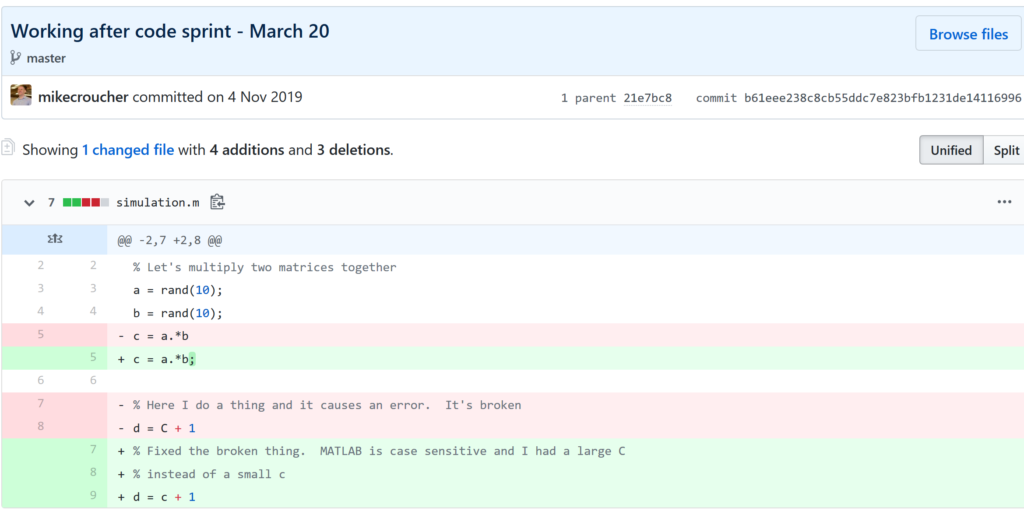
Fixing filename hell
Is this a familiar sight in your working directory?
mycode.py
mycode_jane.py
mycode_ver1b.py
mycode_ver1c.py
mycode_ver1b_january.py
mycode_ver1b_january_BROKEN.py
mycode_ver1b_january_FIXED.py
mycode_ver1b_january_FIXED_for_supervisor.pyFor many people, this is just the beginning. For a project that has existed long enough there might be dozens or even hundreds of these simple scripts that somehow define all of part of your computational workflow. Version control isn’t being used because ‘The code is just a simple script developed by one person’ and yet this situation is already becoming the breeding ground for future problems.
- Which one of these files is the most up to date?
- Which one produced the results in your latest paper or report?
- Which one contains the new work that will lead to your next paper?
- Which ones contain deep flaws that should never be used as part of the research?
- Which ones contain possibly useful ideas that have since been removed from the most recent version?
Applying version control to this situation would lead you to a folder containing just one file
mycode.pyAll of the other versions will still be available via the commit history. Nothing is ever lost and you’ll be able to effectively go back in time to any version of mycode.py you like.
A single point of truth
I’ve even seen folders like the one above passed down generations of PhD students like some sort of family heirloom. I’ve seen labs where multple such folders exist across a dozen machines, each one with a mixture of duplicated and unique files. That is, not only is there a confusing mess of files in a folder but there is a confusing mess of these folders!
This can even be true when only one person is working on a project. Perhaps you have one version of your folder on your University HPC cluster, one on your home laptop and one on your work machine. Perhaps you email zipped versions to yourself from time to time. There are many everyday events that can lead to this state of affairs.
By using a GitHub repository you have a single point of truth for your project. The latest version is there. All old versions are there. All discussion about it is there.
Everything…one place.
The power of this simple idea cannot be overstated. Whenever you (or anyone else) wants to use or continue working on your project, it is always obvious where to go. Never again will you waste several days work only to realise that you weren’t working on the latest version.
Keeping track of everything that changed
The latest version of your analysis or simulation is different from the previous one. Thanks to this, it may now give different results today compared to yesterday. Version control allows you to keep track of everything that changed between two versions. Every line of code you added, deleted or changed is highlighted. Combined with your commit messages where you explain why you made each set of changes, this forms a useful record of the evolution of your project.
It is possible to compare the differences between any two commits, not just two consecutive ones which allows you to track the evolution of your project over time.
Always having a working version of your project
Ever noticed how your collaborator turns up unnanounced just as you are in the middle of hacking on your code. They want you to show them your simulation running but right now its broken! You frantically try some of the other files in your folder but none of them seem to be the version that was working last week when you sent the report that moved your collaborator to come to see you.
If you were using version control you could easily stash your current work, revert to the last good commit and show off your work.
Tracking down what went wrong
You are always changing that script and you test it as much as you can but the fact is that the version from last year is giving correct results in some edge case while your current version is not. There are 100 versions between the two and there’s a lot of code in each version! When did this edge case start to go wrong?
With git you can use git bisect to help you track down which commit started causing the problem which is the first step towards fixing it.
Providing a back up of your project
Try this thought experiment: Your laptop/PC has gone! Fire, theft, dead hard disk or crazed panda attack.
It, and all of it’s contents have vanished forever. How do you feel? What’s running through your mind? If you feel the icy cold fingers of dread crawling up your spine as you realise Everything related to my PhD/project/life’s work is lost then you have made bad life choices. In particular, you made a terrible choice when you neglected to take back ups.
Of course there are many ways to back up a project but if you are using the standard version control workflow, your code is automatically backed up as a matter of course. You don’t have to remember to back things up, back-ups happen as a natural result of your everyday way of doing things.
Making your project easier to find and install
There are dozens of ways to distribute your software to someone else. You could (HORRORS!) email the latest version to a colleuage or you could have a .zip file on your web site and so on.
Each of these methods has a small cognitive load for both recipient and sender. You need to make sure that you remember to update that .zip file on your website and your user needs to find it. I don’t want to talk about the email case, it makes me too sad. If you and your collaborator are emailing code to each other, please stop. Think of the children!
One great thing about using GitHub is that it is a standardised way of obtaining software. When someone asks for your code, you send them the URL of the repo. Assuming that the world is a better place and everyone knows how to use git, you don’t need to do anything else since the repo URL is all they need to get your code. a git clone later and they are in business.
Additionally, you don’t need to worry abut remembering to turn your working directory into a .zip file and uploading it to your website. The code is naturally available for download as part of the standard workflow. No extra thought needed!
In addition to this, some popular computational environments now allow you to install packages directly from GitHub. If, for example, you are following standard good practice for building an R package then a user can install it directly from your GitHub repo from within R using the devtools::install_github() function.
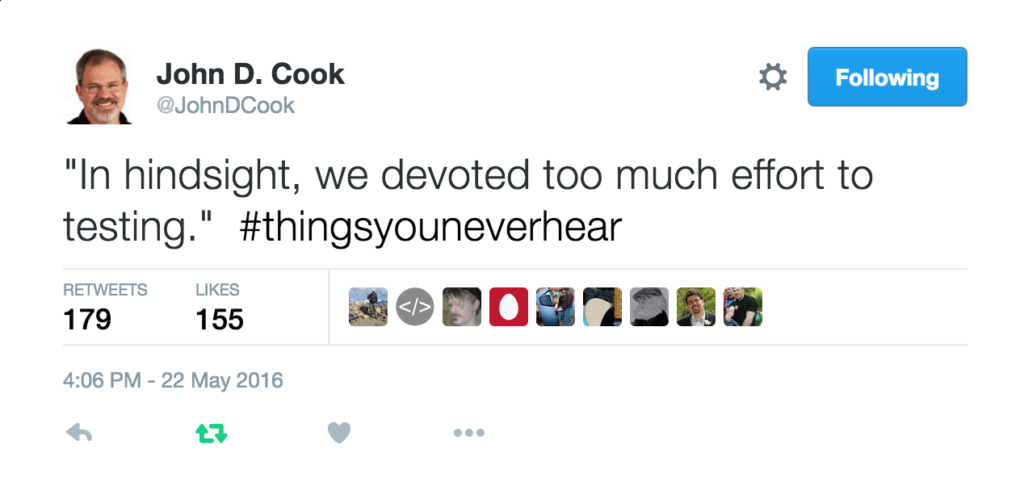
Automatically run all of your tests
You’ve sipped of the KoolAid and you’ve been writing unit tests like a pro. GitHub allows you to link your repo with something called Continuous Integration (CI) that helps maximise the utility of those tests.
Once its all set up the CI service runs every time you, or anyone else, makes a commit to your project. Every time the CI service runs, a virtual machine is created from scratch, your project is installed into it and all of your tests are run with any failures reported.
This gives you increased confidence that everything is OK with your latest version and you can choose to only accept commits that do not break your testing framework.
Collaboration and Community
How git and GitHub can make it easier to collaborate with others on computational projects.
Control exactly who can see your work
‘I don’t want to use GitHub because I want to keep my project private’ is a common reason given to me for not using the service. The ability to create private repositories has been free for some time now (Price plans are available here https://github.com/pricing) and you can have up to 3 collaborators on any of your private repos before you need to start paying. This is probably enough for most small academic projects.
This means that you can control exactly who sees your code. In the early stages it can be just you. At some point you let a couple of trusted collaborators in and when the time is right you can make the repo public so everyone can enjoy and use your work alongside the paper(s) it supports.
Faciliate discussion about your work
Every GitHub repo comes with an Issues section which is effectively a discussion forum for the project. You can use it to keep track of your project To-Do list, bugs, documentation discussions and so on. The issues log can also be integrated with your commit history. This allows you to do things like git commit -m "Improve the foo algorithm according to the discussion in #34" where #34 refers to the Issue discussion where your collaborator pointed out
Allow others to contribute to your work
You have absolute control over external contributions! No one can make any modifications to your project without your explicit say-so.
I start with the above statement because I’ve found that when explaining how easy it is to collaborate on GitHub, the first question is almost always ‘How do I keep control of all of this?’
What happens is that anyone can ‘fork’ your project into their account. That is, they have an independent copy of your work that is clearly linked back to your original. They can happily work away on their copy as much as they like – with no involvement from you. If and when they want to suggest that some of their modifications should go into your original version, they make a ‘Pull Request’.
I emphasised the word ‘Request’ because that’s exactly what it is. You can completely ignore it if you want and your project will remain unchanged. Alternatively you might choose to discuss it with the contributor and make modifications of your own before accepting it. At the other end of the spectrum you might simply say ‘looks cool’ and accept it immediately.
Congratulations, you’ve just found a contributing collaborator.
Reproducible research
How git and GitHub can contribute to improved reproducible research.
Simply making your software available
A paper published without the supporting software and data is (much!) harder to reproduce than one that has both.
Making your software citable
Most modern research cannot be done without some software element. Even if all you did was run a simple statistical test on 20 small samples, your paper has a data and software dependency. Organisations such as the Software Sustainability Institute and the UK Research Software Engineering Association (among many others) have been arguing for many years that such software and data dependencies should be part of the scholarly record alongside the papers that discuss them. That is, they should be archived and referenced with a permanent Digital Object Identifier (DOI).
Once your code is in GitHub, it is straightforward to archive the version that goes with your latest paper and get it its own DOI using services such as Zenodo. Your University may also have its own archival system. For example, The University of Sheffield in the UK has built a system called ORDA which is based on an institutional Figshare instance which allows Sheffield academics to deposit code and data for long term archival.
Which version gave these results?
Anyone who has worked with software long enough knows that simply stating the name of the software you used is often insufficient to ensure that someone else could reproduce your results. To help improve the odds, you should state exactly which version of the software you used and one way to do this is to refer to the git commit hash. Alternatively, you could go one step better and make a GitHub release of the version of your project used for your latest paper, get it a DOI and cite it.
This doesn’t guarentee reproducibility but its a step in the right direction. For extra points, you may consider making the computational environment reproducible too (e.g. all of the dependencies used by your script – Python modules, R packages and so on) using technologies such as Docker, Conda and MRAN but further discussion of these is out of scope for this article.
Building a computational environment based on your repository
Once your project is on GitHub, it is possible to integrate it with many other online services. One such service is mybinder which allows the generation of an executable environment based on the contents of your repository. This makes your code immediately reproducible by anyone, anywhere.
Similar projects are popping up elsewhere such as The Littlest JupyterHub deploy to Azure button which allows you to add a button to your GitHub repo that, when pressed by a user, builds a server in their Azure cloud account complete with your code and a computational environment specified by you along with a JupterHub instance that allows them to run Jupyter notebooks. This allows you to write interactive papers based on your software and data that can be used by anyone.
Complying with funding and journal guidelines
When I started teaching and advocating the use of technologies such as git I used to make a prediction These practices are so obviously good for computational research that they will one day be mandated by journal editors and funding providers. As such, you may as well get ahead of the curve and start using them now before the day comes when your funding is cut off because you don’t. The resulting debate was usually good fun.
My prediction is yet to come true across the board but it is increasingly becoming the case where eyebrows are raised when papers that rely on software are published don’t come with the supporting software and data. Research Software Engineers (RSEs) are increasingly being added to funding review panels and they may be Reviewer 2 for your latest paper submission.
Other uses of git and GitHub for busy academics
It’s not just about code…..
- Build your own websites using GitHub pasges. Every repo can have its own website served directly from GitHub
- Put your presentations on GitHub. I use reveal.js combined with GitHub pages to build and serve my presentations. That way, whenever I turn up at an event to speak I can use whatever computer is plugged into the projector. No more ‘I don’t have the right adaptor’ hell for me.
- Write your next grant proposal. Use Markdown, LaTex or some other git-friendly text format and use git and GitHub to collaboratively write your next grant proposal
The movie below is a visualisation showing how a large H2020 grant proposal called OpenDreamKit was built on GitHub. Can you guess when the deadline was based on the activity?
Further Resources
Further discussions from scientific computing practitioners that discuss using version control as part of a healthy approach to scientific computing
- Good Enough Practices in Scientific Computing –
- Is Your Research Software Correct? – A presentation from Mike Croucher discussing what can go wrong in computational research and what practices can be adopted to do help us do better
- The Turing Way A handbook of good practice in data science brought to you from the Alan Turning Institute
- A guide to reproducible code in ecology and evolution – A handbook from the British Ecological Society that discusses version control as part of general good practice
Learning version control
Convinced? Want to start learning? Let’s begin!
- Git lesson from Software Carpentry – A free, community written tutorial on the basics of git version control
Graphical User Interfaces to git
If you prefer not to use the command line, try these
I was in Stockholm last week to give an invited talk at the Workshop on Nordic Big Biomedical Data for Action. I was representing the Software Sustainability Institute and delivered the latest version of my talk Is Your Research Software Correct? 
It was a great event which introduced me to some nice initiatives going on waaaay up north. Initiatives such as Code Refinery who’s aims align well with those of the UK’s software sustainability Institute. Code refinery was introduced by Radovan Bast — Slide deck at http://cicero.xyz/v2/remark/github/coderefinery/talk-intro/niasc-2016/talk.md/#1
Other talks included the introduction of a scalable, parallel version of BLAST, Big Data Processing for Genomics and Delivering Bioinformatics Software as Virtual Machine images. I also got chance to geek out with some High Performance Computing and Bioinformatics people over interesting Swedish food.
Slides from most of the talks are available at http://www.nordicehealth.se/2016/12/04/workshop-on-nordic-big-biomedical-data-for-action/
One of the great things about being a Research Software Engineer is the diversity of work you can get involved with. I specialise in smaller interventions which means that I can be working with physicists on Monday, engineers on Tuesday, geneticists on Wednesday….you get the idea.
Last month, I got to work with some Ecologists along with Anna Krystalli. We undertook the arduous journey from Sheffield down to Exeter to deliver talks and workshops at a post-conference symposium on reproducibility in science, organised by Malika Ihle and Isabel Winney, at the International Symposium on Behavioural Ecology.
I gave my talk, Is your research software correct?, and also delivered a workshop on using projects and version control using R and RStudio in the Code Cafe style. For the full write up of the day, see the excellent blog post by Anna over at the Mozilla Science Lab blog.
Updates : More resources
- Elevating the status of code in Ecology. Thanks to @katerererena for pointing this one out to me.
- Anna Krystalli’s course material
I learned about entropy as part of my undergraduate Physics education but it turns out that the concept of entropy turns up in many fields including linguistics, themodynamics, information theory, chemistry and artificial intelligence.
As part of Sheffield’s Open Data Science Initiative, computer scientist, Neil Lawrence, has teamed up with linguist, Dagmar Divjak, to organise a cross-faculty discussion meeting on the subject of entropy.
For more details on the day’s events, and to register, see http://opendsi.cc/ed2016/program
I wasted a little time producing the above logo for the event using Mathematica.
Here’s the source code:-
(*consider column one pixel at a time. Invert the pixel if a random number is below some threshold*)
flipbit[col_, prob_] := Module[{result, temp, x},
result = col;
Do[
If[RandomReal[] <= prob,
If[result[[x]] == 1, result[[x]] = 0, result[[x]] = 1];
]
, {x, 1, Length[col]}
];
Return[result]
]
text = "Entropy";
image = Rasterize[Text[Style[text, White, Italic, 190]],
Background -> Black];
imageData = ImageData[Binarize[image]];
const = 1/Dimensions[imageData][[2]]*0.42;
(*Apply flipbit to all columns. Increase probability of flipping as you move along the x-axis*)
logo =
Transpose[
MapIndexed[flipbit[#1, const*#2[[1]]] &, Transpose[imageData]]];
Image[logo]
Finally, I found this quote about entropy that I quite like:
You should call it entropy, for two reasons. In the first place your uncertainty function has been used in statistical mechanics under that name, so it already has a name. In the second place, and more important, no one really knows what entropy really is, so in a debate you will always have the advantage.
John von Neumann to Claude Shannon a name for his new uncertainty function. Source: Wikiquotes
Back in October, I wrote about the Open Data Science events we’ve starting running at the University of Sheffield. These evening events, held at Sheffield’s The Hide are attended by researchers, students and the occasional random who have an interest in data science (collectively referred to as Data Hipsters by some).
At their core, these events are just an excuse for researchers from many disciplines to get together and explore common interests in an informal way and they’ve been a great success.
This month, we’ve gone big and not just in the ‘big data’ sense. We have two free data science events:
- The Data Hide is an evening session 5 – 7pm on December 15th – Tickets at https://www.eventbrite.co.uk/e/the-data-hide-tickets-19895024554
- The big one, Data @ Sheffield is on December 16th and is an all day event
The Sheffield Open Data Science Initiative
The University of Sheffield Open Data Science Initiative (ODSI) is really starting to take off. So what is it?
From the website, the aims of the ODSI are:
- Make new analysis methodologies available as widely and rapidly as possible with as few conditions on their use as possible (see the ML@SITraN group software pages and the local software page).
- Educate our commercial, scientific and medical partners in the use of these latest methodologies (see http://gpss.cc)
- Act to achieve a balance between data sharing for societal benefit and the right of an individual to own their data. (see our summary of our efforts in public understanding and debate)
My role within this initiative is to work on various aspects of research software throughout the University of Sheffield (and beyond!). I am a fellow of the Software Sustainability Institute and you could sum up everything I try to do with their motto Better Software, Better Research.
Join us on October 20th 2015
We have just started a programme of events which aims to bring together a wide variety of people interested in data, machine learning and research software (my favourite part!). The first such event is at The Data Hide on October 2015 at University of Sheffield.
There will be talk on Research Data Management for Computational Science by @ctjacobs_uk as well as lightning talks: What Kind of AI are we Creating? by @lawrennd, Machine Learning for Chemical Simulations by Chris Handley and a demonstration of how great Reveal.js is by me.
This will be followed by food, beer and an opportunity to chat and geek out.
We would be honoured if you would join us.
- Register for a free ticket (Only 40 places currently and they are ‘selling’ out fast!)
- Details of the October 20th event
Would you like to present at a future event?
Contact me to see what we can do together.
Back in December 2014, I learned that I’d be moving from The University of Manchester to The University of Sheffield to do the type of thing I’ve always done which is a combination of research software engineering and research software support.
I’ve been in Sheffield for two months now and am having a blast! There’s so much cool stuff going on here that it makes my head spin a little and the community at Sheffield have welcomed me with open arms. It truly is a wonderful place in which to work.
One of the departments I’ve started working with is The Sheffield Institute for Translation Neuroscience (SITraN). My contributions have been relatively minor so far – A bit of Python coding for a machine learning project called GPy and some code speed-up work in R for Winston Hide and his collaborators. When I hang out in SITraN, I usually sit with the machine learning people and listen in on their conversions about Python, MATLAB, GPUs, C++ and R — it’s essentially Nerdvana for someone like me.
On to the point of this blog post. SITraN now have their own blog called SITraNsmissions where they’ll be discussing various aspects of their work and how it applies the principles of neuroscience to help treat diseases such as Motor Neurone Disease (MND). In the video below, taken from SITraNsmissions first blog post, Professor Pamela Shaw gives an overview of the work that SITraN does.
I recently had the good fortune to be involved in the creation of a European H2020 grant proposal called OpenDreamKit along with an international team from 15 institutions. My own contributions to this proposal were extremely modest and it was my first ever experience of being directly involved in an academic grant proposal. It’s the very first thing I’ve been involved with as part of my new appointment at The University of Sheffield.
Quoting from the proposal:
OpenDreamKit will deliver a flexible toolkit enabling research groups to set up Virtual Research Environments, customised to meet the varied needs of research projects in pure mathematics and applications and supporting the full research life-cycle from exploration, through proof and publication, to archival and sharing of data and code.
One of the many things that’s so great about this proposal is how it was written. Co-ordinated by Nicolas Thiéry, 33 contributors wrote it in LaTeX with version control provided by git and github. The video below, produced using gource, is a visualation of the github repo over time and shows how we all danced around and with each other. My new manager, Neil Lawrence, who was much more deeply involved than I has good things to say about the process too.
The proposal was submitted yesterday after a lot of hard work and, as Nicholas Thiery commented in one of his emails to the group, is “Open from start to end :-)”
The Sage Facebook page summed up my thoughts about this project perfectly: “See the collaboration behind the *proposal*, and imagine the collaboration in the software!”